Submission overview
What data can be submitted?
ArrayExpress accepts all functional genomics data generated from microarray or next-generation sequencing (NGS) platforms. Popular experiment types are transcription profiling (mRNA and miRNA), SNP genotyping, chromatin immunoprecipitation (ChIP) and comparative genomic hybridisation. Here is the full list of experiment types in ArrayExpress.
For NGS submissions, raw data files are brokered to the European Nucleotide Archive (ENA), while ArrayExpress archives any processed data. Read more about the special rules applying to sequencing submissions.
We currently don't accept metagenomics/metatranscriptomics data (please submit to the EBI Metagenomics service) or de novo transcriptome assembly data (the raw RNA-seq reads should be submitted to ArrayExpress, but the assembled transcriptome file directly to ENA).
ArrayExpress accepts all functional genomics data generated from microarray or next-generation sequencing (NGS) platforms. Popular experiment types are transcription profiling (mRNA and miRNA), SNP genotyping, chromatin immunoprecipitation (ChIP) and comparative genomic hybridisation. Here is the full list of experiment types in ArrayExpress.
For NGS submissions, raw data files are brokered to the European Nucleotide Archive (ENA), while ArrayExpress archives any processed data. Read more about the special rules applying to sequencing submissions.
We currently don't accept metagenomics/metatranscriptomics data (please submit to the EBI Metagenomics service) or de novo transcriptome assembly data (the raw RNA-seq reads should be submitted to ArrayExpress, but the assembled transcriptome file directly to ENA).
What do I need to prepare?
Breadth of the data and metadata
The aim is that an ArrayExpress user should have everything they need for the data set to make sense and be reproducible without referring to an associated paper.
Microarray submissions follow the "Minimum Information About a Microarray Experiment" (MIAME) guidelines. Sequencing submissions follow a similar set of guidelines, "Minimum Information About a Sequencing Experiment" (MINSEQE).
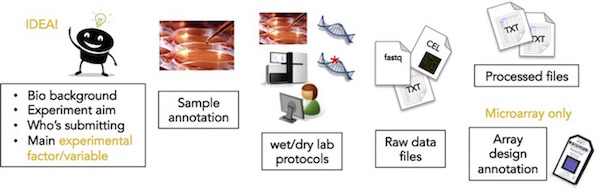
As a submitter, you may need to consult with your colleagues, e.g. collaborators or the core facility personnel performing microarray hybridisation or sequencing for you, to gather all detailed information for a successful submission.
Metadata
- Experiment description to give context to the data set (not pasting publication abstract please)
- Protocols of all experimental (e.g. sample sourcing, sequencing library preparation) and data analysis procedures
- Sample annotation (as much details as possible, e.g. age of the plant, mouse strain, cell type)
- Author information (please include the principal investigator of the project)
- Sequencing library specification (NGS experiments only)
Raw data
- Unprocessed data files
Microarray: files obtained from the microarray scanner (e.g. Affymetrix CEL files, Agilent feature extraction txt files, Illumina idat files)
See the full list of accepted microarray raw data files.Sequencing: compressed raw sequence read files (e.g. fastq.gz files)
See accepted sequencing raw data files.
Processed data
- Processed data matrix in tab-delimited txt format
- Other processed data formats, e.g. bam alignment files (optional)