Array design submissions
What is an array design?
For a microarray experiment, an array design is the unique set of probes found on the microarray chip. In ArrayExpress, an ADF (Array Design Format) file captures information about a microarray chip. The ADF is a spreadsheet-like tab-delimited text file with some metadata header rows, followed by a multi-column table of probe information (here is an example). Data from a microarray experiment cannot be interpreted properly if the array design used is unclear, especially when the microarray is custom-made for a particular research project.
Do I need to submit an array design for my experiment?
Many of the array designs, especially commercially available ones, are already accessioned in ArrayExpress as ADF files (one ADF for one microarray chip), in which case you do not need to submit any ADF to us. All you need to do is to specify the ADF by accession when asked by Annotare "What array did you use?".
To find out whether your array design is already in ArrayExpress and get its accession, you can search for array designs in the BioStudies browser. Go to the ArrayExpress collection and write the type:"array" in the search box. Then add any keywords (e.g. manufacturer name, species name) after "AND", e.g. find all human arrays with this query. If you know the accession of the array design ("platform") from the NCBI GEO database, you can convert it to the ArrayExpress equivalent by replacing "GPL" with "A-GEOD", e.g. "GPL1234" will be "A-GEOD-1234". If you cannot find a GEO platform accession in ArrayExpress, email us, and we will import the array design.
How do I submit an array designs?
1. Download an array design file (ADF) template
Pick a template that suits your microarray. (Check out this concepts page if you are unsure whether your microarray contains features, reporters or composite elements.)
- Commercially available arrays or custom arrays from commercial manufacturers (e.g. NimbleGen, Agilent, Affymetrix, Illumina), just metadata header to fill in, and send us any supporting files*: commercial array design header template
- Custom spotted arrays with features in block row / block column / row / column grid coordinates: spotted array template
- Custom arrays with only reporters (probes), without grid coordinate information: reporter array template
- Custom arrays with reporters and composite elements: composite array template
*Supporting files are those from the microarray's manufacturer containing information of each probe, e.g. NDF files for NimbleGen arrays, GAL files for spotted arrays. An ArrayExpress curator will convert your supporting file into the ADF table, and append it to the metadata header you fill in to create a complete, valid ADF.
2. Fill in template with array design information
Refer to this ADF creation help page for an explanation on how to fill in the template (e.g. what is mandatory, what are accepted values). Please stick to the guidelines so your ADF can be processed promptly. All the ADF header fields and ADF table column headings, highlighted in bold, are from a controlled vocabulary, so please do not change them.
When you've finished filling in the template, please save the file in tab-delimited text (*.txt) format. (If you're unsure about this format, this is how you do it with Microsoft Excel or OpenOffice Calc.)
Below is an example snippet of a filled template (text in light blue filled in by the submitter):
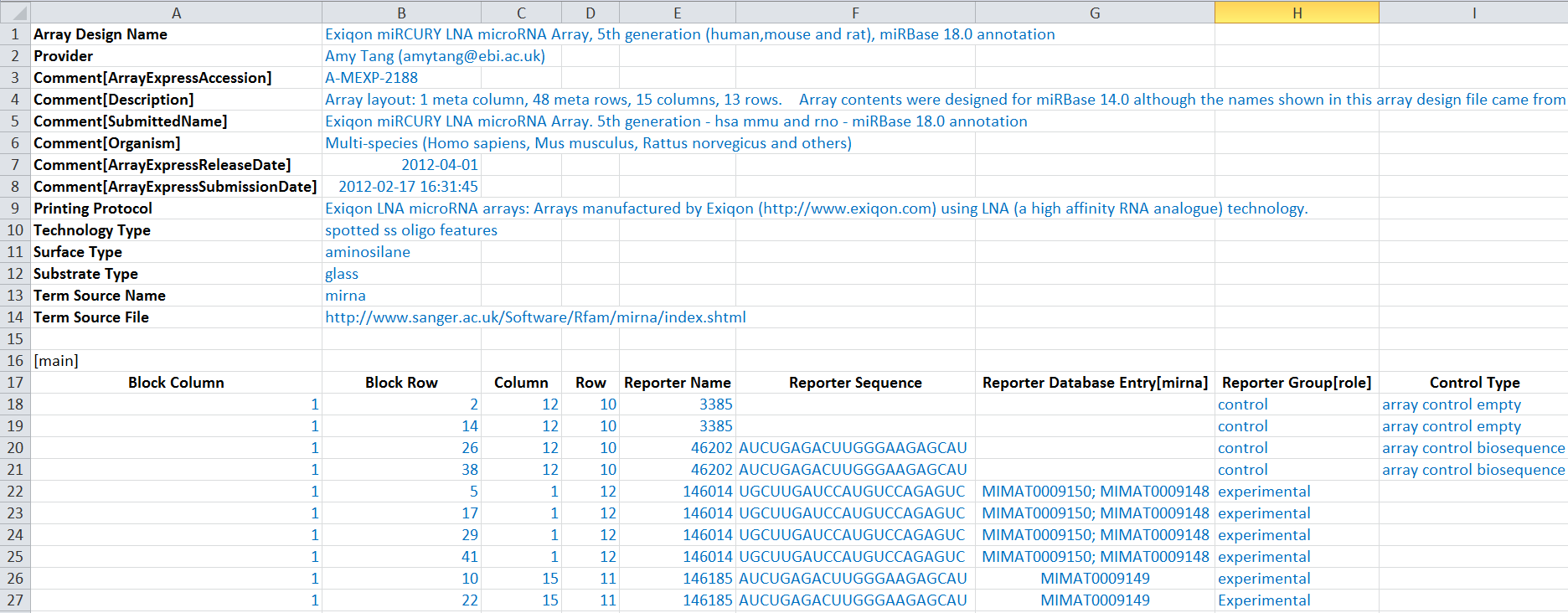
3. Send the completed ADF file via FTP and then email us
Please send the ADF file by FTP/Aspera (instructions here). If you are submitting a commercially available array design, don't forget to also send us the supporting files (e.g. NDF file for NimbleGen arrays, GAL file for spotted arrays).
When the FTP/Aspera transfer is completed, please email us at annotare@ebi.ac.uk with the name of the transferred file(s).
An ArrayExpress curator will get in touch by email once he/she has looked at your ADF file, including any questions or comments he/she may have. For commercial array designs, the curator will create the full ADF on your behalf by combining the metadata header and some information converted from the annotation/support files.